Unlocking opioid neuropeptide dynamics with genetically-encoded biosensors Nat Neurosci.
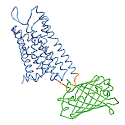
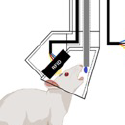
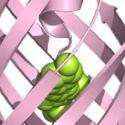
Endogenous peptides are central to addiction research, but there are limited experimental tools to study these systems in a circuit-specific and selective spatiotemporal manner. Tian and collaborators developed a class of genetically encoded opioid peptide indicators, κLight, δLight, and µLight, based on κOR, δOR, and µOR respectively, that are selective and sensitive to the rapid dynamic changes in endogenous opioid peptides release in vivo.
[Read More]