Nanoparticle single-cell multiomic readouts reveal that cell heterogeneity influences lipid nanoparticle-mediated messenger RNA delivery
Published in Nature Nanotechnology.
Authors
Curtis Dobrowolski, Kalina Paunovska, Elisa Schrader Echeverri, David Loughrey, Alejandro J. Da Silva Sanchez, Huanzhen Ni, Marine Z. C. Hatit, Melissa P. Lokugamage, Yanina Kuzminich, Hannah E. Peck, Philip J. Santangelo & James E. Dahlman
Paper presented by Dr. Atul Daiwile and selected by the NIDA TDI Paper of the Month Committee
Background and Technological Advancement
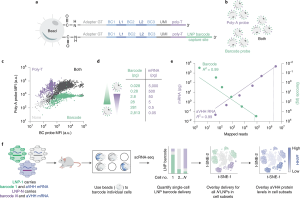
Cells within tissues can be described as homogeneous but are composed of subsets of cells with distinct transcriptional states that may influence their ability to undergo gene transfer by nanoparticles. To test this hypothesis, researchers from the Georgia Institute of Technology and Emory University’s School of Medicine have developed a system that makes nanoparticle delivery to subsets of cells more predictable by combining DNA barcoding with a nanoparticle delivery system called single-cell nanoparticle targeting-sequencing (SENT-seq). Using SENT-seq, the investigators identified how different nanoparticles deliver DNA barcodes and mRNA into cells to affect the transcriptome and proteome with single cell resolution. They also identified cell subtypes that exhibit relatively high and low nanoparticle uptake in a heterogeneous cell population which has not been previously possible using conventional techniques.
Nanoparticle single-cell multiomic readouts reveal that cell heterogeneity influences lipid nanoparticle-mediated messenger RNA delivery Journal Article
In: Nat Nanotechnol, vol. 17, no. 8, pp. 871–879, 2022, ISSN: 1748-3395.