Image copyright – Science.
Dense functional and molecular readout of a circuit hub in sensory cortex
Published in Science.
Authors
Cameron Condylis, Abed Ghanbari, Nikita Manjrekar, Karina Bistrong, Shenqin Yao, Zizhen Yao, Thuc Nghi Nguyen, Hongkui Zeng, Bosiljka Tasic, and Jerry L. Chen
Paper presented by Dr. Atul Daiwile and selected by the NIDA TDI Paper of the Month Committee.
Background and Technological Advancement
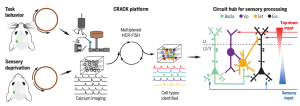
The molecular signatures (e.g. gene expression profile) of cells in the brain are used to identify the cellular components of neuronal circuitry. To identify cell types involved in neuronal circuits associated with a specific behavior, Dr. Chen’s lab from Boston University developed a platform designated “Comprehensive readout of activity and cell type markers (CRACK)”. The CRACK platform combines multi-area two photon calcium imaging microscopy with hybridization chain reaction–fluorescence in situ hybridization (HCR-FISH) to label and track mRNA. CRACK allows researchers to first observe the electrical firing of neurons in the brain of a live mouse during a behavioral task, and then track the expression of specific genes in slices of the animal’s brain, ultimately linking specific cells and their molecular signatures to particular behaviors. Now with CRACK, researchers can record activity of multiple neurons and precisely identify those neurons. Through simultaneous imaging across identified cell types, they were able to measure functional connectivity between subpopulations of neurons for a behavioral task.
Dense functional and molecular readout of a circuit hub in sensory cortex Journal Article
In: Science, vol. 375, no. 6576, pp. eabl5981, 2022, ISSN: 1095-9203.
