Image copyright – Nature.
Transgenic mice for in vivo epigenome editing with CRISPR-based systems
Published in Nature Methods.
Authors
Matthew P. Gemberling, Keith Siklenka, Erica Rodriguez, Katherine R. Tonn-Eisinger, Alejandro Barrera, Fang Liu, Ariel Kantor, Liqing Li, Valentina Cigliola, Mariah F. Hazlett, Courtney A. Williams, Luke C. Bartelt, Victoria J. Madigan, Josephine C. Bodle, Heather Daniels, Douglas C. Rouse, Isaac B. Hilton, Aravind Asokan, Maria Ciofani, Kenneth D. Poss, Timothy E. Reddy, Anne E. West & Charles A. Gersbach
Paper presented by Dr. Katherine Savell and selected by the NIDA TDI Paper of the Month Committee
Background and Technological Advancement
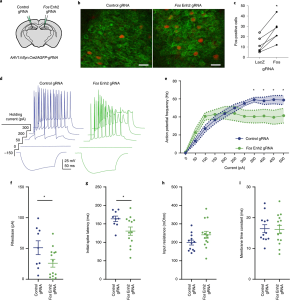
Advances in CRISPR-based tools offer gene-specific control of transcriptional and epigenetic states. However, achieving expression in targeted cell types and tissues is challenging due to the size of dCas9-effector fusion proteins, particularly in the central nervous system. Gemberling et al. generate and validate of two new Cre-dependent transgenic mouse lines harboring LSL-dCas9-p300 (gene activator/histone acetyltransferase) and LSL-dCas9-KRAB (gene repressor/histone methyltransferase). To achieve cell specific gene activation or repression in the brain, a gene-specific guide RNA is delivered by viral vector and cells expressing a cell-specific Cre will activate or repress the target gene of interest. The authors demonstrate that guide RNAs targeting gene enhancers and the gene promoter are effective at modulating both histone modification state and gene transcription. The two new transgenic mouse lines can be paired with any Cre-driver mouse for bidirectional modulation of gene expression in specific cell types.
Transgenic mice for in vivo epigenome editing with CRISPR-based systems Journal Article
In: Nat Methods, vol. 18, no. 8, pp. 965–974, 2021, ISSN: 1548-7105.
