Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging
Published in Cell.
 Authors
Authors
Peggie Cheung, Francesco Vallania, Hayley C Warsinske, Michele Donato, Steven Schaffert, Sarah E Chang, Mai Dvorak, Cornelia L Dekker, Mark M Davis, Paul J Utz, Purvesh Khatri, and Alex J Kuo
Paper presented by Dr. Atul Daiwile and selected by the NIDA TDI Paper of the Month Committee
Publication Brief Description
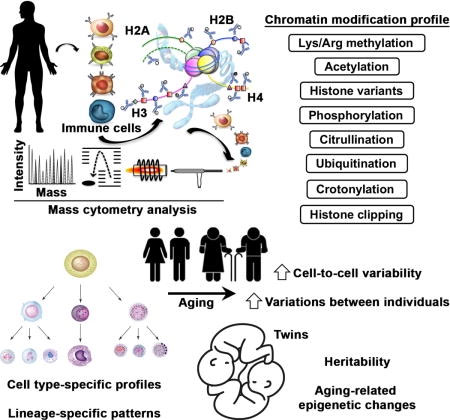
Post-translational modifications of histone proteins are central to the regulation of nearly all DNA-based biological processes. However, the degree and variability of chromatin modifications in specific human cells remain largely unknown. A group of researchers from Stanford University employed a multiplexed mass cytometry to profile the epigenetic landscape and measure a broad array of global chromatin modifications in human cells at the single-cell level. Their data revealed a marked difference in cell type and hematopoietic-lineage-specific chromatin modification patterns. Using the “EpiTOF” method they observed an age-related increase in the heterogeneity between individuals and elevated cell-to-cell variability in chromatin modifications Moreover, they discovered that aging-related chromatin alterations are predominantly driven by non-heritable influences. The EpiTOF technology provides new opportunities for identifying cell-specific epigenetic changes associated with altered physiological and pathological states.
Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging Journal Article
In: Cell, vol. 173, no. 6, pp. 1385–1397.e14, 2018, ISSN: 1097-4172.
