Image copyright – Neuron.
Enhancer viruses for combinatorial cell-subclass-specific labeling
Published in Neuron.
Authors: Lucas T Graybuck, Tanya L Daigle, Adriana E Sedeño-Cortés, Miranda Walker, Brian Kalmbach, Garreck H Lenz, Elyse Morin, Thuc Nghi Nguyen, Emma Garren, Jacqueline L Bendrick, Tae Kyung Kim, Thomas Zhou, Marty Mortrud, Shenqin Yao, La’ Akea Siverts, Rachael Larsen, Bryan B Gore, Eric R Szelenyi, Cameron Trader, Pooja Balaram, Cindy T J van Velthoven, Megan Chiang, John K Mich, Nick Dee, Jeff Goldy, Ali H Cetin, Kimberly Smith, Sharon W Way, Luke Esposito, Zizhen Yao, Viviana Gradinaru, Susan M Sunkin, Ed Lein, Boaz P Levi, Jonathan T Ting, Hongkui Zeng, Bosiljka Tasic
Paper presented by Dr. Katherine Savell and selected by the NIDA TDI Paper of the Month Committee
Publication Brief Description
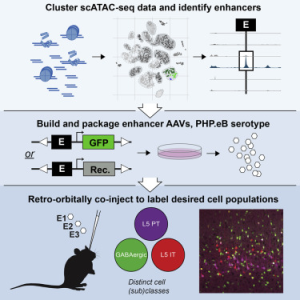
The brain is comprised of multiple regions of heterogeneous cell types. Experimental access to distinct cell types is necessary to study normal function and disease states of the brain, and it is essential to determine if defined cell types are conserved between species. Two studies published last year harnessed single-cell chromatin accessibility to define and test cell-type-distinguishing enhancers for generating AAVs to express transgenes in discrete cell populations, and to study expression correspondence between species. These approaches provide researchers with one avenue to access specific cell types in the brain for targeted population studies and gene therapies.
Enhancer viruses for combinatorial cell-subclass-specific labeling Journal Article
In: Neuron, vol. 109, no. 9, pp. 1449–1464.e13, 2021, ISSN: 1097-4199.
