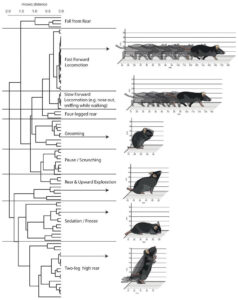
Image copyright Nature Neuroscience.
Revealing the structure of pharmacobehavioral space through motion sequencing
Published in Nature Neuroscience (2020)
Authors
Alexander B. Wiltschko, Tatsuya Tsukahara, Ayman Zeine, Rockwell Anyoha, Winthrop F. Gillis, Jeffrey E. Markowitz, Ralph E. Peterson, Jesse Katon, Matthew J. Johnson, and Sandeep Robert Datta
Paper presented by Dr. Thomas Stalnaker and selected by the NIDA TDI Paper of the Month Committee.
Publication Brief Description
Standard behavioral assays that are used for screening candidate therapeutic drugs, such as the elevated plus maze and forced swim test, yield only one or two dimensions of information. Wiltschko et al. used a machine learning algorithm called MoSeq (for motion sequencing) to analyze 3D video recordings of twenty minutes of open field mouse activity, thereby obtaining a much higher dimensional readout of behavior in response to a wide variety of psychoactive drugs and doses. They show that MoSeq can distinguish almost all tested drugs and doses better than standard analyses of behavior. In a proof of principle, they go on to show that MoSeq can identify on- and off-target effects of candidate therapeutic drugs in a mouse model of autism. The MoSeq tool, which is readily accessible, could potentially be used to screen anti-addiction drugs and might also be useful to analyze behavioral changes in models of drug relapse or in models of drug or non-drug decision making studied at NIDA.
Revealing the structure of pharmacobehavioral space through motion sequencing. Journal Article
In: Nat Neurosci., vol. 23, iss. 11, pp. 1433-1443, 2020.
