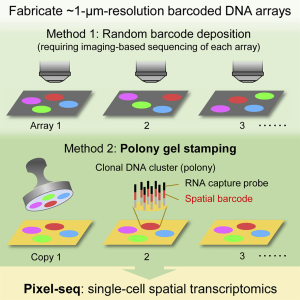
Polony gels enable amplifiable DNA stamping and spatial transcriptomics of chronic pain
Published in Cell.
Authors
Xiaonan Fu, Li Sun, Runze Dong, Jane Y Chen, Runglawan Silakit, Logan F Condon, Yiing Lin, Shin Lin, Richard D Palmiter, Liangcai Gu
Paper presented by Dr. Katherine Savell and selected by the NIDA TDI Paper of the Month Committee
Background and Technological Advancement
Advances in spatial transcriptomics offer transcriptional characterization of complex tissues, such as the brain. However, most existing methods only provide a spatial resolution of 100-200 µm, with gene expression representing multiple cells in proximity. Higher resolution is possible but requires specialized equipment and expensive reagents. Fu*, Sun*, Dong* et. al. generated and validated a novel polony-based ‘stamp gel’ that uses common lab equipment and enzymatic replication to produce copies of the 1 µm resolution barcoded array slide that is used for spatial transcriptomic capture. The authors demonstrate that the stamping fidelity is robust and consistent over many cycles, and the slides capture the top layer of RNA on a tissue slice. The authors created a new bioinformatics tool called “V-seg” to assign transcripts to single cells which they used to investigate the effects of chronic pain on cell-type transcriptomes in the parabrachial nucleus. Taken together, polony gel stamping is poised to increase accessibility of high-resolution spatial transcriptomics.
Polony gels enable amplifiable DNA stamping and spatial transcriptomics of chronic pain Journal Article
In: Cell, vol. 185, no. 24, pp. 4621–4633.e17, 2022, ISSN: 1097-4172.