Reviews To Read – May 2017
Stolzenberg, Sebastian; Michino, Mayako; LeVine, Michael V; Weinstein, Harel; Shi, Lei
Computational approaches to detect allosteric pathways in transmembrane molecular machines. Journal Article
In: Biochim Biophys Acta, vol. 1858, no. 7 Pt B, pp. 1652-1662, 2016, ISSN: 0006-3002 (Print); 0006-3002 (Linking).
@article{Stolzenberg:2016aa,
title = {Computational approaches to detect allosteric pathways in transmembrane molecular machines.},
author = {Sebastian Stolzenberg and Mayako Michino and Michael V LeVine and Harel Weinstein and Lei Shi},
url = {https://www.ncbi.nlm.nih.gov/pubmed/26806157},
doi = {10.1016/j.bbamem.2016.01.010},
issn = {0006-3002 (Print); 0006-3002 (Linking)},
year = {2016},
date = {2016-07-01},
journal = {Biochim Biophys Acta},
volume = {1858},
number = {7 Pt B},
pages = {1652-1662},
address = {Computational Molecular Biology Group, Institute for Mathematics, Arnimallee 6, 14195, Berlin, Germany; Department of Physiology and Biophysics, Weill Medical College of Cornell University, New York, NY 10065, USA; Department of Physics, Cornell University, Ithaca, NY 14850, USA.},
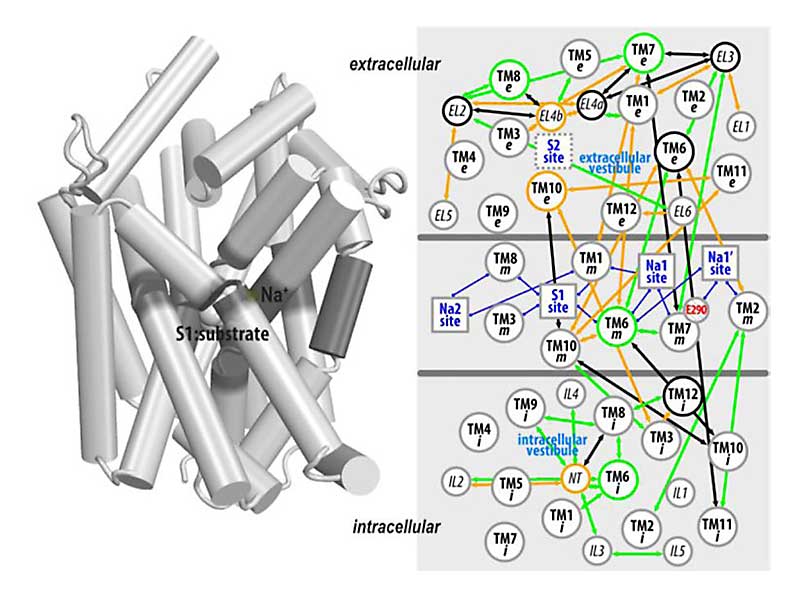
abstract = {Many of the functions of transmembrane proteins involved in signal processing and transduction across the cell membrane are determined by allosteric couplings that propagate the functional effects well beyond the original site of activation. Data gathered from breakthroughs in biochemistry, crystallography, and single molecule fluorescence have established a rich basis of information for the study of molecular mechanisms in the allosteric couplings of such transmembrane proteins. The mechanistic details of these couplings, many of which have therapeutic implications, however, have only become accessible in synergy with molecular modeling and simulations. Here, we review some recent computational approaches that analyze allosteric coupling networks (ACNs) in transmembrane proteins, and in particular the recently developed Protein Interaction Analyzer (PIA) designed to study ACNs in the structural ensembles sampled by molecular dynamics simulations. The power of these computational approaches in interrogating the functional mechanisms of transmembrane proteins is illustrated with selected examples of recent experimental and computational studies pursued synergistically in the investigation of secondary active transporters and GPCRs. This article is part of a Special Issue entitled: Membrane Proteins edited by J.C. Gumbart and Sergei Noskov.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Many of the functions of transmembrane proteins involved in signal processing and transduction across the cell membrane are determined by allosteric couplings that propagate the functional effects well beyond the original site of activation. Data gathered from breakthroughs in biochemistry, crystallography, and single molecule fluorescence have established a rich basis of information for the study of molecular mechanisms in the allosteric couplings of such transmembrane proteins. The mechanistic details of these couplings, many of which have therapeutic implications, however, have only become accessible in synergy with molecular modeling and simulations. Here, we review some recent computational approaches that analyze allosteric coupling networks (ACNs) in transmembrane proteins, and in particular the recently developed Protein Interaction Analyzer (PIA) designed to study ACNs in the structural ensembles sampled by molecular dynamics simulations. The power of these computational approaches in interrogating the functional mechanisms of transmembrane proteins is illustrated with selected examples of recent experimental and computational studies pursued synergistically in the investigation of secondary active transporters and GPCRs. This article is part of a Special Issue entitled: Membrane Proteins edited by J.C. Gumbart and Sergei Noskov.