Reviews To Read – January 2024.
Published in Neuroscience & Biobehavioral Reviews by Khalid Elhadi, Atul P. Daiwile, and Jean Lud Cadet of the NIDA IRP Molecular Neuropsychiatry Research Section.
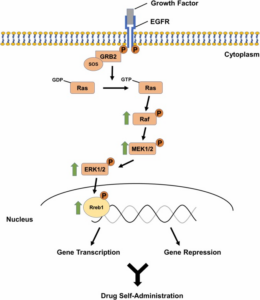
Methamphetamine use disorder (MUD) is very widespread in the world because methamphetamine is easy to make and cheap to buy. Heavy users usually take the drug in binges, try to stop on their own or in a treatment program, but often suffer relapses. We and others are trying to identify the basis for MUD by using animal models. We have used different schedules of methamphetamine self-administration to identify epigenetic modifications and changes in gene and protein expression in different brain regions that we think are involved in substance use disorders (SUDs). We have found that there are lots of changes in the rat dorsal striatum following methamphetamine intake. Specifically, we found changes in the expression of genes that regulate transcription, potassium channel function, and neuroinflammation. Our studies suggest the need for more research in this area because there are still no medications approved by the FDA for MUD.
In: Neuroscience & Biobehavioral Reviews, vol. 155, pp. 105440, 2023, ISSN: 0149-7634.