Background | Status & Availability | Transgene Info | Phenotypic Characterization | Breeding | Genotyping | References | Blog/Comments/Reviews | Related rats | Acknowledgements
Background
Fluorescent proteins are routinely used to label and identify cells mammalian cells. mCherry is a red fluorescent protein with an excitation/emission max of 587/610 (Clontech/Takara). Here, we created a transgenic rat with a Cre-dependent mCherry transgene expressed from the EF1a promoter. We show that this rat can be used in combination with viral vector-mediated delivery of Cre to produce mCherry expression in the rat brain or crossed with transgenic rats expressing Cre-recombinase to selectively label neurons such as the dopaminergic neurons in the midbrain.
Status and Availability
This rat has been published (PMID: 30792150).
As of April 22, 2016, this strain is available as line#00687 at RRRC.![]()
This rat is registered at the Rat Genome Database (RGD) as RGD ID#8693598.![]()
Transgene Information
Figure 1. Schematic of the DIO-mCherry transgene. The insert encoding mCherry was amplified with linkered oligos and Addgene #26975 as a template. This insert was recombined into the backbone (pAAV EF1a DIO EYFP, Addgene 27056, digested with NheI and AscI restriction enzymes) using In-Fusion cloning mix (Clontech) to produce pOTTC337 (Addgene #47626). The pOTTC337 (pAAV EF1a DIO mCherry) plasmid was digested with MluI and RsrII, gel purified away from the plasmid backbone, and microinjected into fertilized oocytes harvested from a Long Evans rat by the NIMH Transgenic Core. Surviving pups were screened for the integrated transgene by PCR genotyping. This line (LE-Tg(DIO-mCherry)2Ottc) has 0.5-0.7 copies of the transgene per copy of Ggt1 (indicating 1 copy per haploid genome) as determined by droplet digital PCR.
Phenotypic Characterization
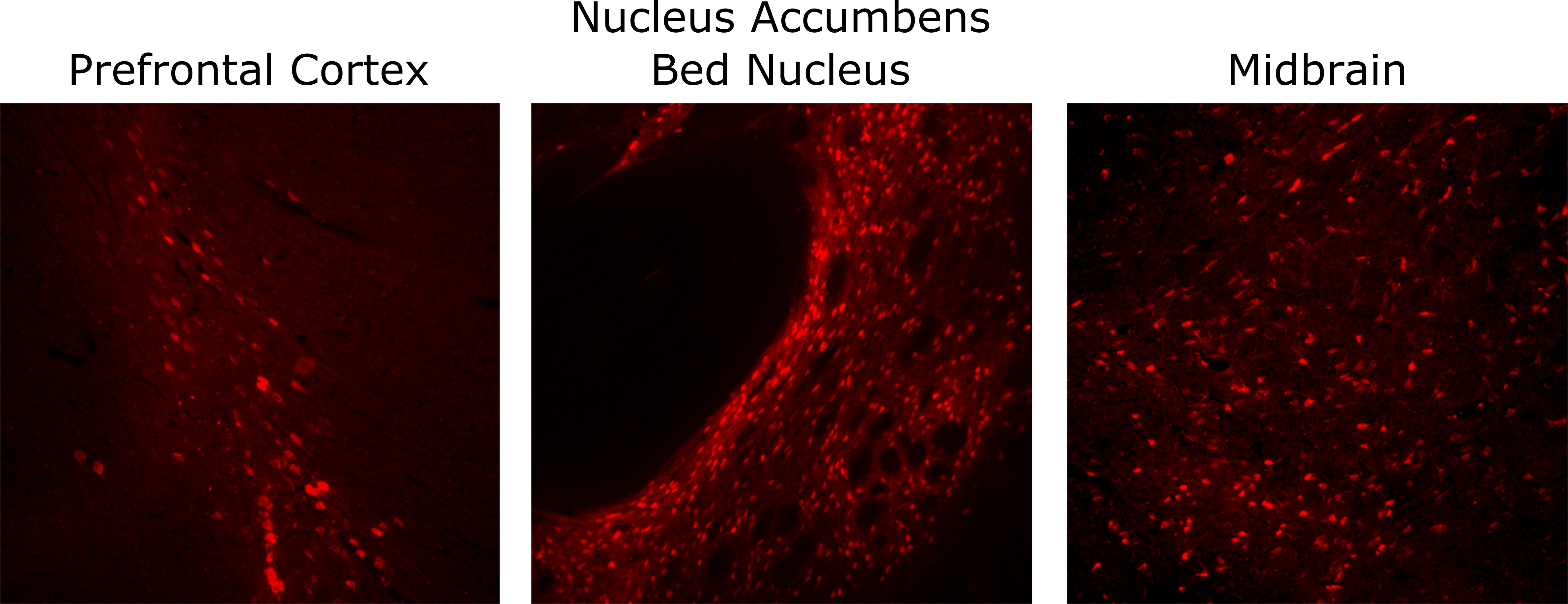 Figure 2 Legend: AAV-Cre injections into different brain regions of LE-Tg(DIO-mCherry)2Ottc. A 12 week old, male DIO-mCherry rat was injected with AAV1-Cre virus into different brain regions using following coordinates relative to bregma: prefrontal cortex (AP=+2.5 mm, ML = 0.8 mm, DV=-3.5 mm), nucleus accumbens/Bed Nucleus (AP=+1.5 mm, ML = 1.4 mm, DV=-7.5 mm) and midbrain (AP=-6.0 mm, ML = 1.9 mm, DV=-7.2). Five weeks after injection, the animal was perfused with 4% PFA and cryosections (40 um thickness) were used for imaging by epifluorescent microscopy. Representative images of mCherry fluorescence are shown (red). Similar fluorescent patterns were not observed in uninjected, contralateral hemispheres. Download print resolution version here.
Figure 2 Legend: AAV-Cre injections into different brain regions of LE-Tg(DIO-mCherry)2Ottc. A 12 week old, male DIO-mCherry rat was injected with AAV1-Cre virus into different brain regions using following coordinates relative to bregma: prefrontal cortex (AP=+2.5 mm, ML = 0.8 mm, DV=-3.5 mm), nucleus accumbens/Bed Nucleus (AP=+1.5 mm, ML = 1.4 mm, DV=-7.5 mm) and midbrain (AP=-6.0 mm, ML = 1.9 mm, DV=-7.2). Five weeks after injection, the animal was perfused with 4% PFA and cryosections (40 um thickness) were used for imaging by epifluorescent microscopy. Representative images of mCherry fluorescence are shown (red). Similar fluorescent patterns were not observed in uninjected, contralateral hemispheres. Download print resolution version here.
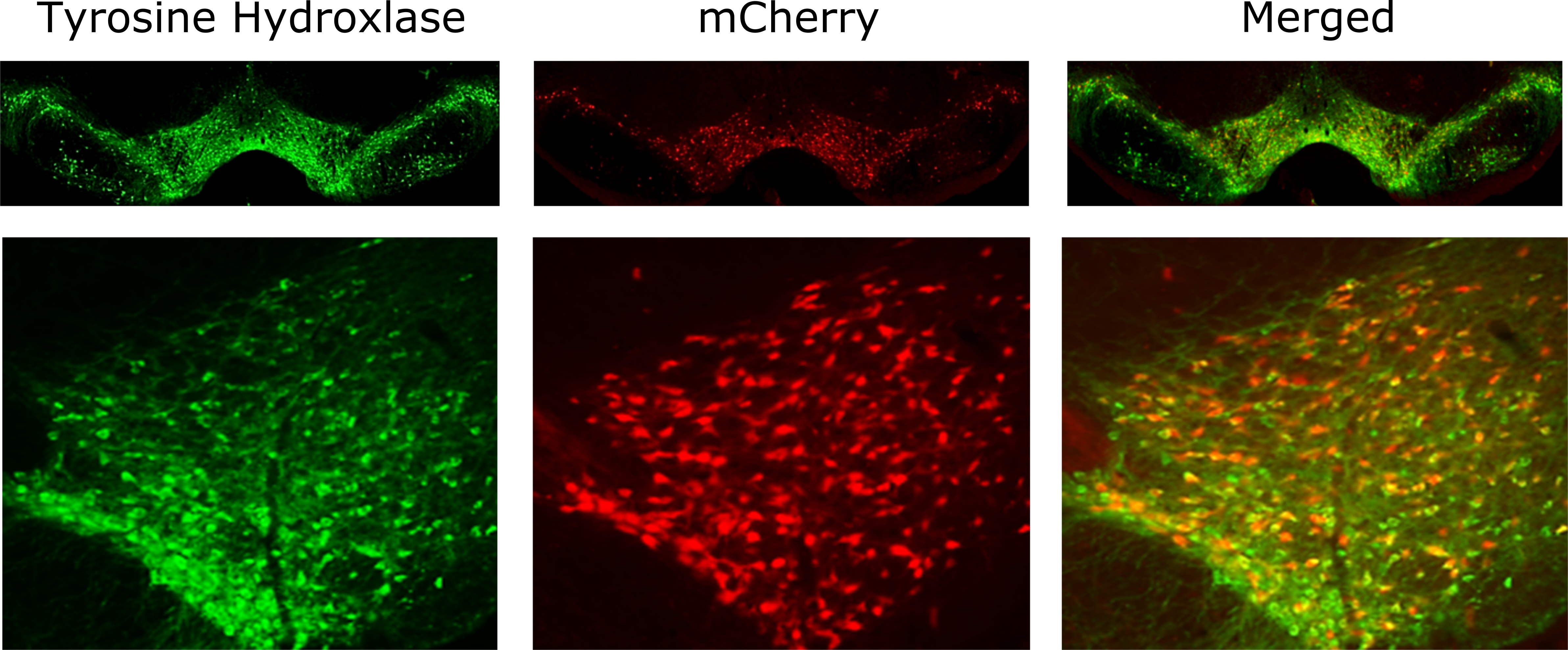 Figure 3 Legend: Midbrain images of progeny from LE-Tg(DAT-iCre)6Ottc rats crossed with LE-Tg(DIO-mCherry)2Ottc. Midbrain sections from adult female rat produced from the cross of DAT-Cre and DIO-mCherry parents. Cryosections (40 um thickness) were immunostained for tyrosine hydroxylase (TH) to identify dopaminergic neurons. Sections were imaged using epifluorescence to detect the TH immunoreactivity (green) and mCherry (red). Top row shows low magnification of both hemispheres of midbrain. Bottom row is higher magnification of VTA region showing approximately >99% of mCherry cells colocalizing with TH. Download print resolution version here.
Figure 3 Legend: Midbrain images of progeny from LE-Tg(DAT-iCre)6Ottc rats crossed with LE-Tg(DIO-mCherry)2Ottc. Midbrain sections from adult female rat produced from the cross of DAT-Cre and DIO-mCherry parents. Cryosections (40 um thickness) were immunostained for tyrosine hydroxylase (TH) to identify dopaminergic neurons. Sections were imaged using epifluorescence to detect the TH immunoreactivity (green) and mCherry (red). Top row shows low magnification of both hemispheres of midbrain. Bottom row is higher magnification of VTA region showing approximately >99% of mCherry cells colocalizing with TH. Download print resolution version here.
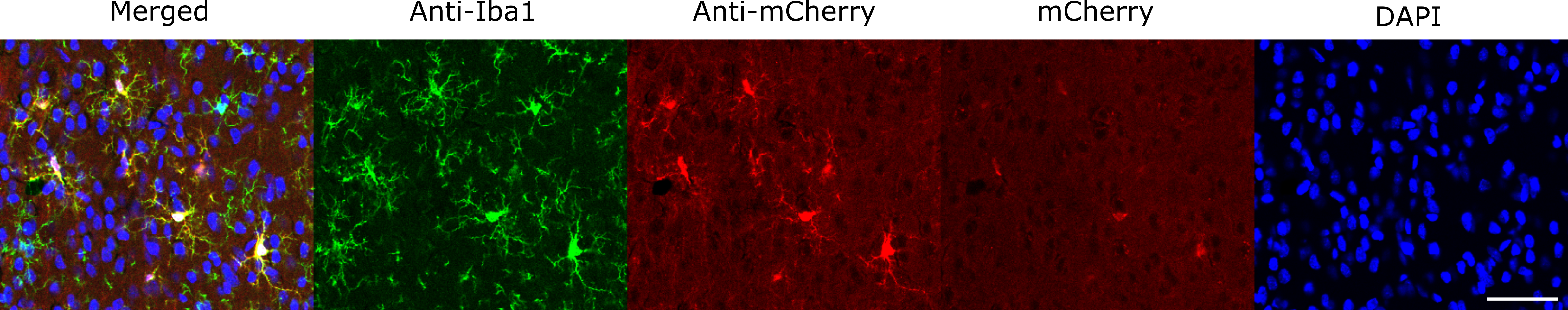 Figure 4 Legend: Prefrontal cortex images of progeny from LE-Tg(CX3CR1-CreERT2)3Ottc rats crossed with LE-Tg(DIO-mCherry)2Ottc. Prefrontal cortex sections from 10-week old male rat produced from the cross of CX3CR1-CreERT2 and DIO-mCherry parents injected with tamoxifen at P10. Cryosections were immunostained for Iba1 (Waco Cat# 019-19741) to identify microglia and anti-mCherry (Takara Cat# 632543) to enhance the detection of weak mCherry expression. Sections were imaged using epifluorescence to detect the Iba1 immunoreactivity (green), mCherry immunoreactivity (red), and DAPI (blue). Scale bar represents 50 microns. Download print resolution version here.
Figure 4 Legend: Prefrontal cortex images of progeny from LE-Tg(CX3CR1-CreERT2)3Ottc rats crossed with LE-Tg(DIO-mCherry)2Ottc. Prefrontal cortex sections from 10-week old male rat produced from the cross of CX3CR1-CreERT2 and DIO-mCherry parents injected with tamoxifen at P10. Cryosections were immunostained for Iba1 (Waco Cat# 019-19741) to identify microglia and anti-mCherry (Takara Cat# 632543) to enhance the detection of weak mCherry expression. Sections were imaged using epifluorescence to detect the Iba1 immunoreactivity (green), mCherry immunoreactivity (red), and DAPI (blue). Scale bar represents 50 microns. Download print resolution version here.
Breeding Strategy
Breeding Information, click here for PDF
Genotyping Assays
Click here for PDF
References that cite this rat
2019
Neuron-Specific Genome Modification in the Adult Rat Brain Using CRISPR-Cas9 Transgenic Rats. Journal Article
In: Neuron, vol. 102, no. 1, pp. 105–119, 2019, ISSN: 1097-4199 (Electronic); 0896-6273 (Linking).
Blog/Comments/Reviews
Last Updated on November 12, 2024
No comments or reviews are available at this time.
Other related rats
No related rats at this time.
DIO-mCherry can be crossed with other Cre-expressing lines found on the OTTC – Transgenic Rats list.
Acknowledgements
YaJun Zhang, Julie Necarsulmer, Chris Richie, Brandon Harvey, Janette Lebron

