Hot Off the Press! – April 2016
Cadet, J L; Brannock, C; Krasnova, I N; Jayanthi, S; Ladenheim, B; McCoy, M T; Walther, D; Godino, A; Pirooznia, M; Lee, R S
In: Mol Psychiatry, vol. 22, no. 8, pp. 1196–1204, 2016, ISSN: 1476-5578 (Electronic); 1359-4184 (Linking).
@article{Cadet2017,
title = {Genome-wide DNA hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence.},
author = {J L Cadet and C Brannock and I N Krasnova and S Jayanthi and B Ladenheim and M T McCoy and D Walther and A Godino and M Pirooznia and R S Lee},
url = {http://www.ncbi.nlm.nih.gov/pubmed/27046646},
doi = {10.1038/mp.2016.48},
issn = {1476-5578 (Electronic); 1359-4184 (Linking)},
year = {2016},
date = {2016-04-05},
journal = {Mol Psychiatry},
volume = {22},
number = {8},
pages = {1196--1204},
address = {Molecular Neuropsychiatry Research Branch, NIDA Intramural Research Program, Baltimore, MD, USA.},
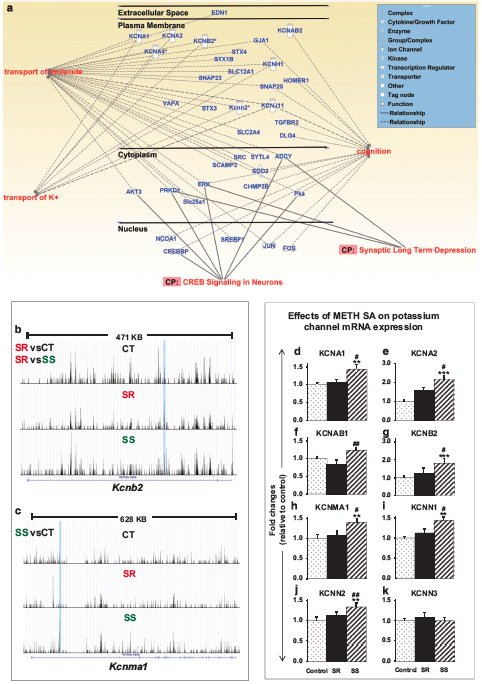
abstract = {Epigenetic consequences of exposure to psychostimulants are substantial but the relationship of these changes to compulsive drug taking and abstinence is not clear. Here, we used a paradigm that helped to segregate rats that reduce or stop their methamphetamine (METH) intake (nonaddicted) from those that continue to take the drug compulsively (addicted) in the presence of footshocks. We used that model to investigate potential alterations in global DNA hydroxymethylation in the nucleus accumbens (NAc) because neuroplastic changes in the NAc may participate in the development and maintenance of drug-taking behaviors. We found that METH-addicted rats did indeed show differential DNA hydroxymethylation in comparison with both control and nonaddicted rats. Nonaddicted rats also showed differences from control rats. Differential DNA hydroxymethylation observed in addicted rats occurred mostly at intergenic sites located on long and short interspersed elements. Interestingly, differentially hydroxymethylated regions in genes encoding voltage (Kv1.1, Kv1.2, Kvb1 and Kv2.2)- and calcium (Kcnma1, Kcnn1 and Kcnn2)-gated potassium channels observed in the NAc of nonaddicted rats were accompanied by increased mRNA levels of these potassium channels when compared with mRNA expression in METH-addicted rats. These observations indicate that changes in differentially hydroxymethylated regions and increased expression of specific potassium channels in the NAc may promote abstinence from drug-taking behaviors. Thus, activation of specific subclasses of voltage- and/or calcium-gated potassium channels may provide an important approach to the beneficial treatment for METH addiction.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Epigenetic consequences of exposure to psychostimulants are substantial but the relationship of these changes to compulsive drug taking and abstinence is not clear. Here, we used a paradigm that helped to segregate rats that reduce or stop their methamphetamine (METH) intake (nonaddicted) from those that continue to take the drug compulsively (addicted) in the presence of footshocks. We used that model to investigate potential alterations in global DNA hydroxymethylation in the nucleus accumbens (NAc) because neuroplastic changes in the NAc may participate in the development and maintenance of drug-taking behaviors. We found that METH-addicted rats did indeed show differential DNA hydroxymethylation in comparison with both control and nonaddicted rats. Nonaddicted rats also showed differences from control rats. Differential DNA hydroxymethylation observed in addicted rats occurred mostly at intergenic sites located on long and short interspersed elements. Interestingly, differentially hydroxymethylated regions in genes encoding voltage (Kv1.1, Kv1.2, Kvb1 and Kv2.2)- and calcium (Kcnma1, Kcnn1 and Kcnn2)-gated potassium channels observed in the NAc of nonaddicted rats were accompanied by increased mRNA levels of these potassium channels when compared with mRNA expression in METH-addicted rats. These observations indicate that changes in differentially hydroxymethylated regions and increased expression of specific potassium channels in the NAc may promote abstinence from drug-taking behaviors. Thus, activation of specific subclasses of voltage- and/or calcium-gated potassium channels may provide an important approach to the beneficial treatment for METH addiction.